import shap
import matplotlib.pyplot as plt
%matplotlib inline
import numpy as np
import pandas as pd
from xgboost import XGBRegressor
# Import machine learning methods
from sklearn.model_selection import train_test_split
from sklearn import datasets
from sklearn.metrics import mean_absolute_error, mean_absolute_percentage_error, \
r2_score, root_mean_squared_error
# Import shap for shapley values
import shap
# JavaScript Important for the interactive charts later on
shap.initjs()16 SHAP with regression trees (Diabetes Progression Dataset)
From the SKLearn documentation:
Number of Instances: 442
Number of Attributes: First 10 columns are numeric predictive values
Target: Column 11 is a quantitative measure of disease progression one year after baseline
Attribute Information:
age age in years
sex
bmi body mass index
bp average blood pressure
s1 tc, total serum cholesterol
s2 ldl, low-density lipoproteins
s3 hdl, high-density lipoproteins
s4 tch, total cholesterol / HDL
s5 ltg, possibly log of serum triglycerides level
s6 glu, blood sugar level
https://scikit-learn.org/stable/datasets/toy_dataset.html#diabetes-dataset
Note: Each of these 10 feature variables have been mean centered and scaled by the standard deviation times the square root of n_samples (i.e. the sum of squares of each column totals 1). This isn’t necessary given we’ve opted to use a tree model, but will be fine for the purpose of demonstration.
# Load the diabetes dataset
X, y = datasets.load_diabetes(return_X_y=True, as_frame=True, scaled=False)
X.head()| age | sex | bmi | bp | s1 | s2 | s3 | s4 | s5 | s6 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 59.0 | 2.0 | 32.1 | 101.0 | 157.0 | 93.2 | 38.0 | 4.0 | 4.8598 | 87.0 |
| 1 | 48.0 | 1.0 | 21.6 | 87.0 | 183.0 | 103.2 | 70.0 | 3.0 | 3.8918 | 69.0 |
| 2 | 72.0 | 2.0 | 30.5 | 93.0 | 156.0 | 93.6 | 41.0 | 4.0 | 4.6728 | 85.0 |
| 3 | 24.0 | 1.0 | 25.3 | 84.0 | 198.0 | 131.4 | 40.0 | 5.0 | 4.8903 | 89.0 |
| 4 | 50.0 | 1.0 | 23.0 | 101.0 | 192.0 | 125.4 | 52.0 | 4.0 | 4.2905 | 80.0 |
# let's just rename these to be clearer based on the data dictionary
X = X.rename(columns=
{
"s1": "total_serum_cholesterol",
"s2": "ldl_cholesterol",
"s3": "hdl_cholesterol",
"s4": "total_cholesterol",
"s5": "serum_triglycerides_log",
"s6": "blood_sugar"
})y.head()0 151.0
1 75.0
2 141.0
3 206.0
4 135.0
Name: target, dtype: float64
# Split the data into training/testing sets
diabetes_X_train, diabetes_X_test, diabetes_y_train, diabetes_y_test = train_test_split(
X, y,
test_size = 0.25,
random_state=42
)
model = XGBRegressor(random_state=42)
# Train the model using the training sets
model.fit(diabetes_X_train, diabetes_y_train)
# Make predictions using the testing set
diabetes_y_pred = model.predict(diabetes_X_test)
print(f"Mean absolute error: {mean_absolute_error(diabetes_y_test, diabetes_y_pred):.2f}")
print(f"Mean absolute percentage error: {mean_absolute_percentage_error(diabetes_y_test, diabetes_y_pred):.2%}" )
print("Root Mean squared error: %.2f" % root_mean_squared_error(diabetes_y_test, diabetes_y_pred))
# The coefficient of determination: 1 is perfect prediction
print("Coefficient of determination: %.2f" % r2_score(diabetes_y_test, diabetes_y_pred))Mean absolute error: 46.59
Mean absolute percentage error: 39.84%
Root Mean squared error: 58.01
Coefficient of determination: 0.39# explain the model's predictions using SHAP
explainer = shap.Explainer(model, diabetes_X_train)
shap_values = explainer(diabetes_X_test)
shap_values.values =
array([[ 10.93827235, 2.8060772 , -1.00666967, ..., -4.88568904,
12.19191076, 3.60045137],
[ -3.45617191, 3.4392566 , 4.56441772, ..., 0.37978809,
-13.22084821, 13.30567497],
[ 10.71647202, -1.89627643, -4.11494224, ..., -1.75186097,
31.18730879, -0.99276986],
...,
[ 2.63871242, 4.10990574, 21.90833779, ..., 0.12410758,
33.50004507, 4.06646104],
[ -0.19391851, 7.1562762 , 21.04501401, ..., 0.28448169,
-46.11341172, 8.68782261],
[ 8.6260937 , -5.57821297, -20.22131193, ..., -3.46062915,
6.32432399, -1.93388196]])
.base_values =
array([167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748, 167.10061748,
167.10061748, 167.10061748, 167.10061748])
.data =
array([[ 61. , 1. , 25.8 , ..., 5. , 4.9972, 90. ],
[ 74. , 1. , 29.8 , ..., 3. , 4.3944, 86. ],
[ 66. , 2. , 26. , ..., 4. , 5.5683, 87. ],
...,
[ 55. , 1. , 28.2 , ..., 4. , 5.366 , 103. ],
[ 53. , 1. , 28.8 , ..., 3.15 , 4.0775, 85. ],
[ 67. , 2. , 23. , ..., 5. , 4.654 , 99. ]])shap_values_numeric = shap_values.values
shap_values_numericarray([[ 10.93827235, 2.8060772 , -1.00666967, ..., -4.88568904,
12.19191076, 3.60045137],
[ -3.45617191, 3.4392566 , 4.56441772, ..., 0.37978809,
-13.22084821, 13.30567497],
[ 10.71647202, -1.89627643, -4.11494224, ..., -1.75186097,
31.18730879, -0.99276986],
...,
[ 2.63871242, 4.10990574, 21.90833779, ..., 0.12410758,
33.50004507, 4.06646104],
[ -0.19391851, 7.1562762 , 21.04501401, ..., 0.28448169,
-46.11341172, 8.68782261],
[ 8.6260937 , -5.57821297, -20.22131193, ..., -3.46062915,
6.32432399, -1.93388196]])17 Plots
# visualize the first prediction's explanation
shap.plots.waterfall(shap_values[0])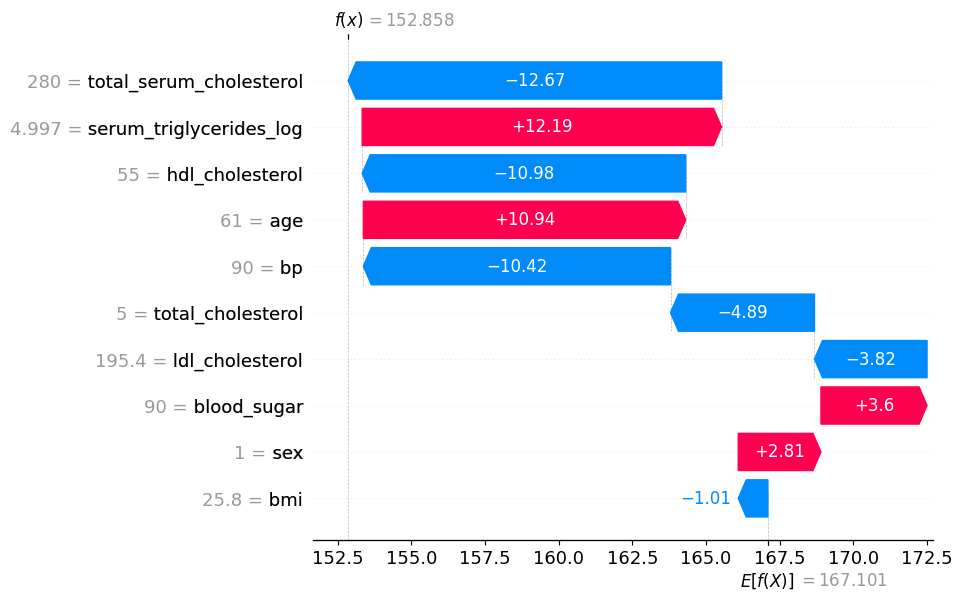
# visualize a later prediction's explanation
shap.plots.waterfall(shap_values[7])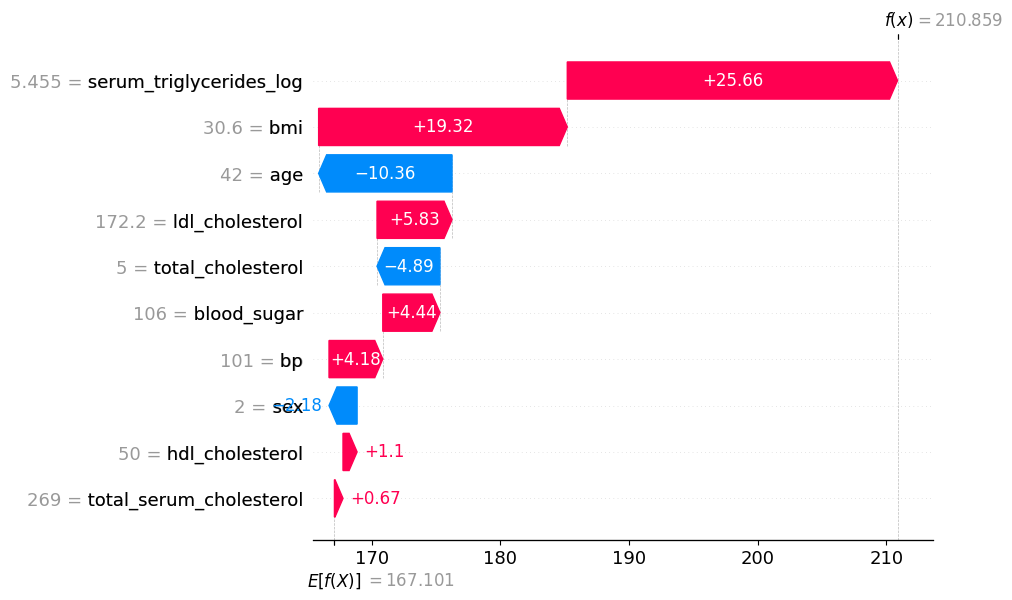
17.1 Force plots
# visualize the first prediction's explanation with a force plot
shap.plots.force(shap_values[0])Have you run `initjs()` in this notebook? If this notebook was from another user you must also trust this notebook (File -> Trust notebook). If you are viewing this notebook on github the Javascript has been stripped for security. If you are using JupyterLab this error is because a JupyterLab extension has not yet been written.
# visualize all the predictions
shap.plots.force(shap_values)Have you run `initjs()` in this notebook? If this notebook was from another user you must also trust this notebook (File -> Trust notebook). If you are viewing this notebook on github the Javascript has been stripped for security. If you are using JupyterLab this error is because a JupyterLab extension has not yet been written.
17.2 Dependence Plots
# create a dependence scatter plot to show the effect of a single feature across the whole dataset
shap.plots.scatter(shap_values[:, "age"])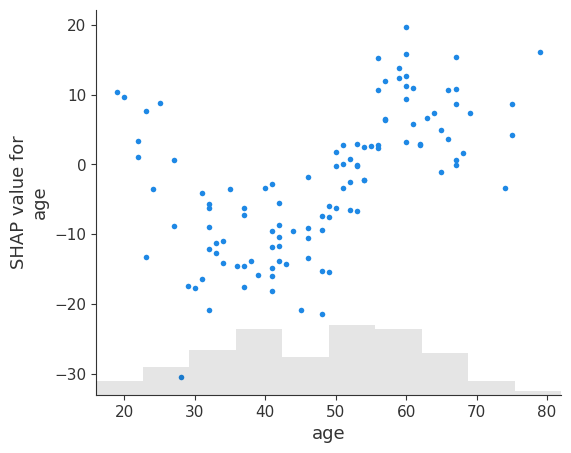
# create a dependence scatter plot to show the effect of a single feature across the whole dataset
shap.plots.scatter(shap_values[:, "bmi"], color=shap_values)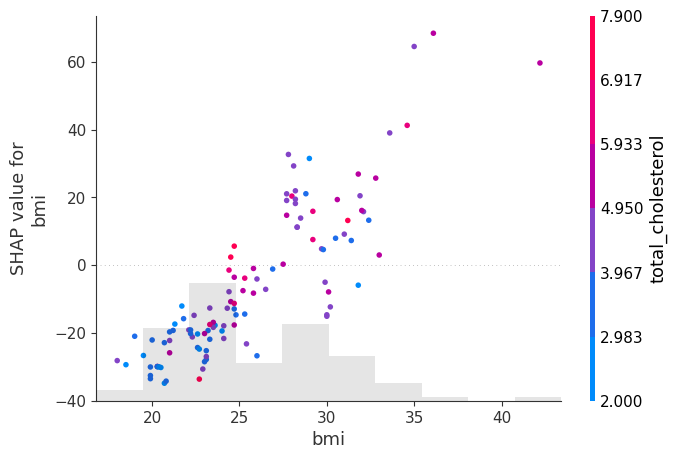
# create a dependence scatter plot to show the effect of a single feature across the whole dataset
shap.plots.scatter(shap_values[:, "blood_sugar"], color=shap_values)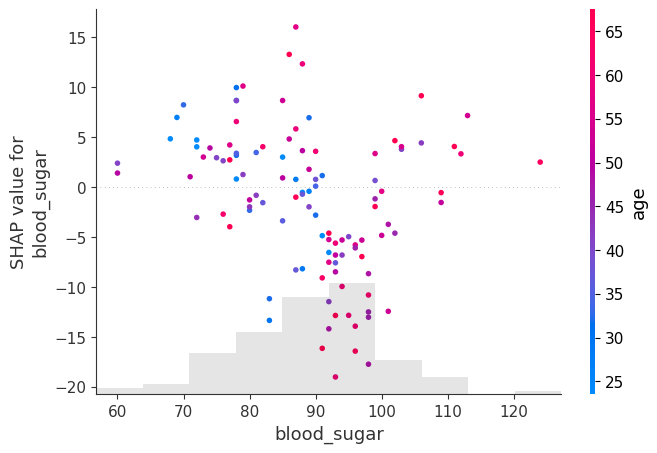
shap.plots.scatter(shap_values[:, "age"], color=shap_values[:, "bmi"])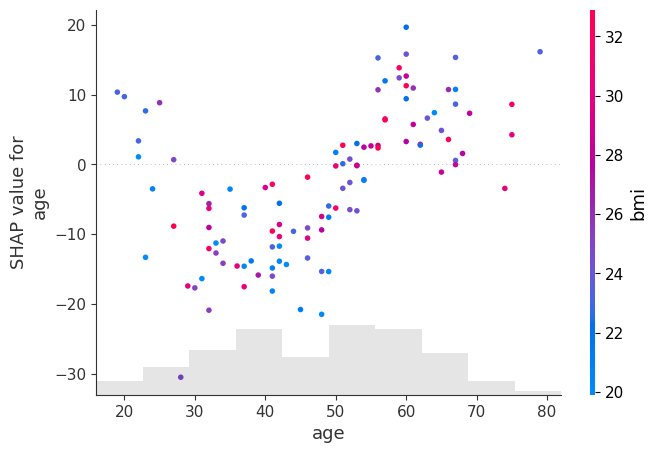
shap.plots.scatter(shap_values[:, "bmi"], color=shap_values[:, "age"])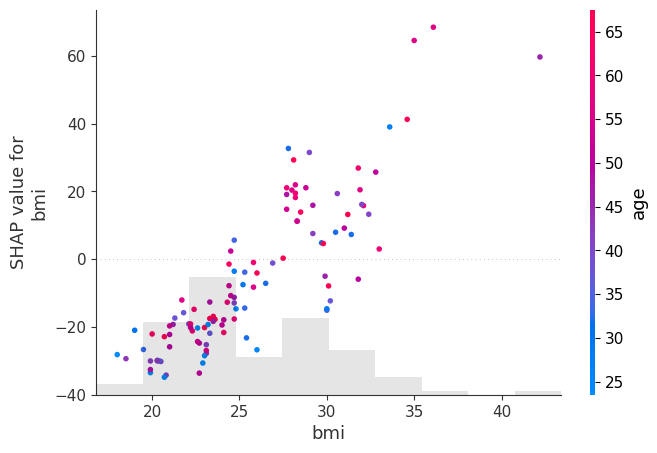
shap.plots.scatter(shap_values[:, "age"], color=shap_values[:, "bmi"])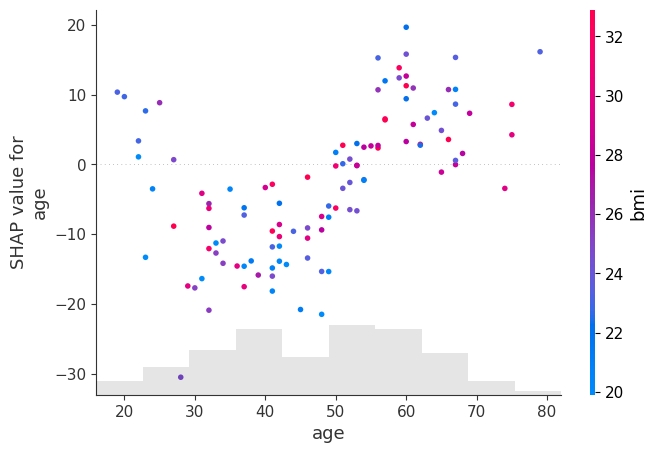
17.3 Beeswarm
# summarize the effects of all the features
shap.plots.beeswarm(shap_values)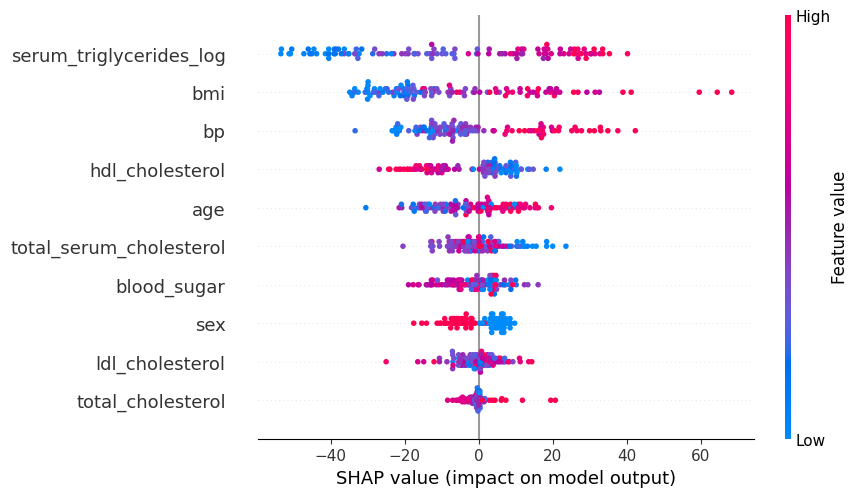
17.4 Violin
# summarize the effects of all the features
shap.plots.violin(shap_values)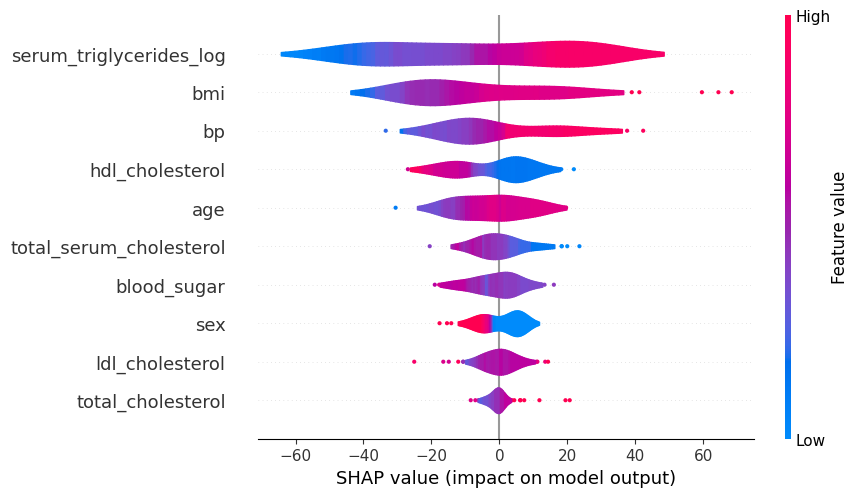
17.5 Bar
shap.plots.bar(shap_values)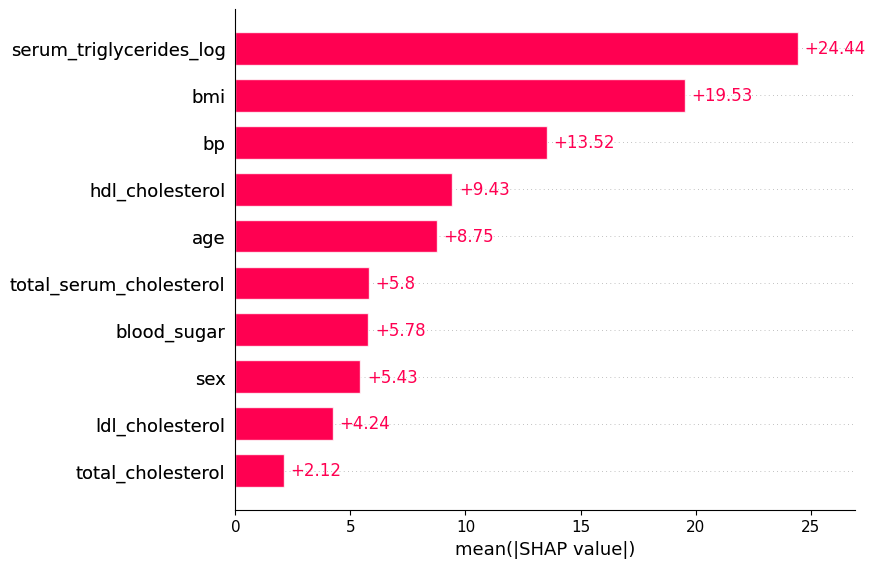
17.5.1 Splitting by cohorts
bmi_category = ["<30" if shap_values[i, "bmi"].data < 30 else ">=30" for i in range(shap_values.shape[0])]
shap.plots.bar(shap_values.cohorts(bmi_category).abs.mean(0))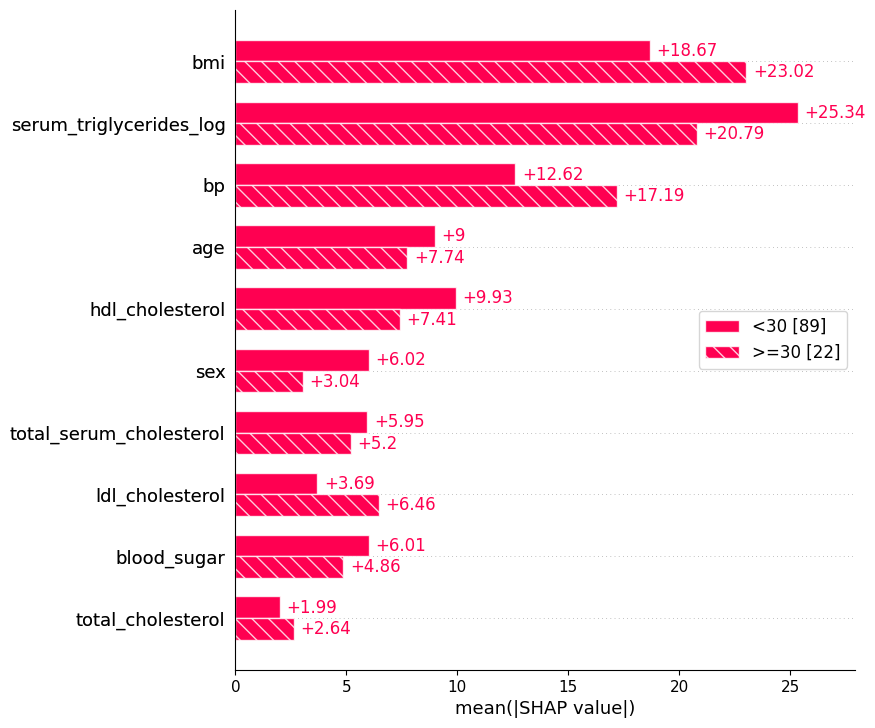
17.5.2 Automatic cohort splitting
shap.plots.bar(shap_values.cohorts(2).abs.mean(0))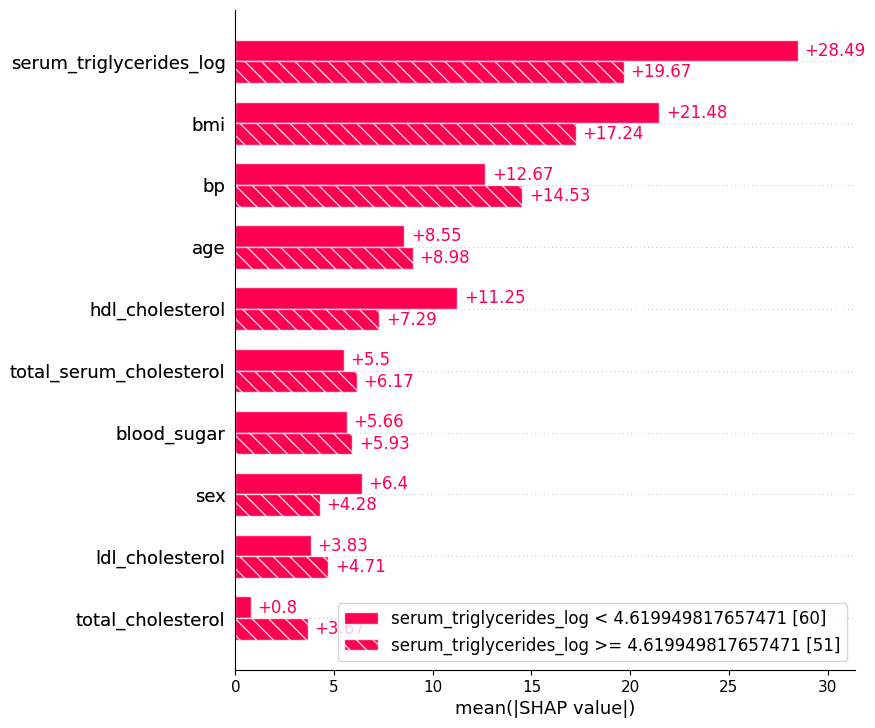
Plot the bars for an individual.
shap.plots.bar(shap_values[0])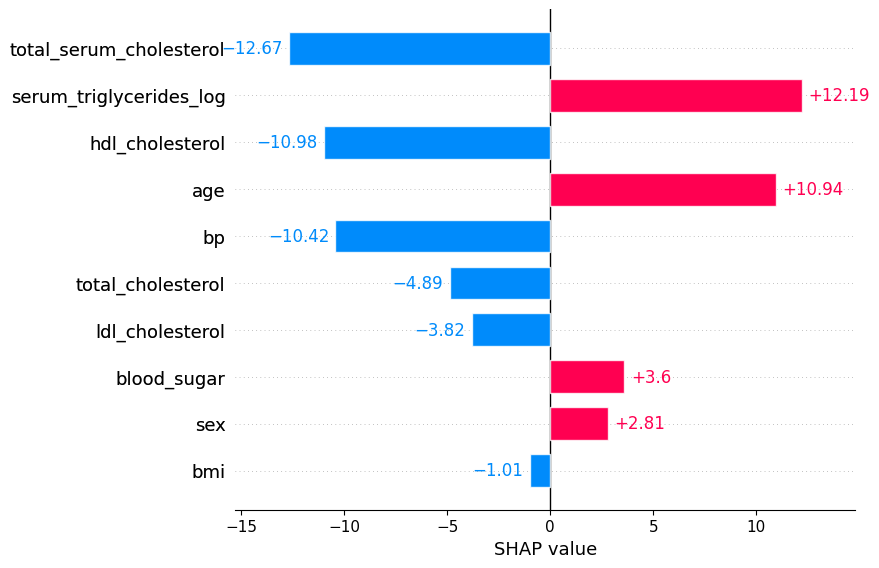
17.6 Heatmap
shap.plots.heatmap(shap_values)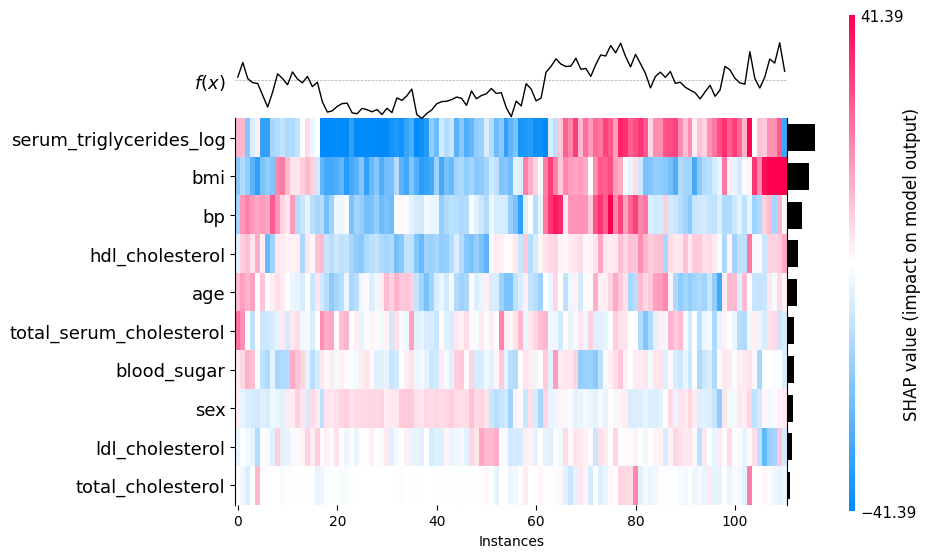
17.7 Decision
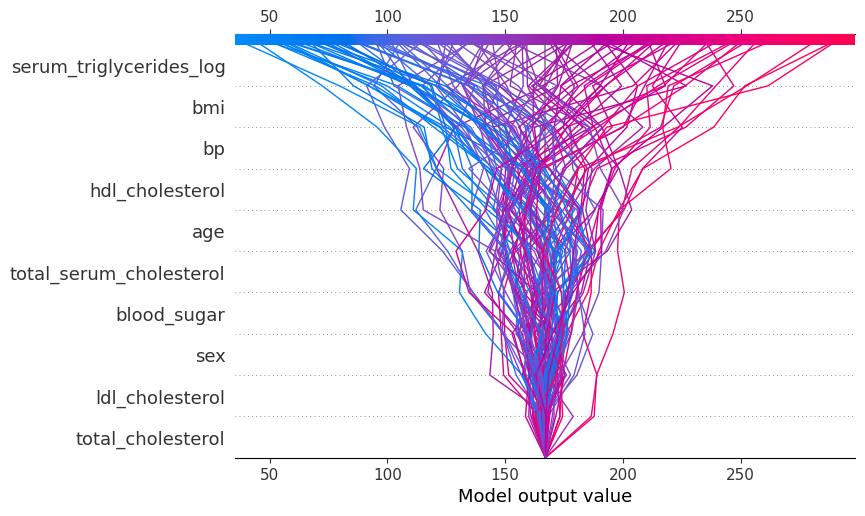
shap.plots.decision(
explainer.expected_value,
explainer.shap_values(diabetes_X_test),
feature_names=X.columns.tolist()
)
17.7.1 Explanation plot for individual
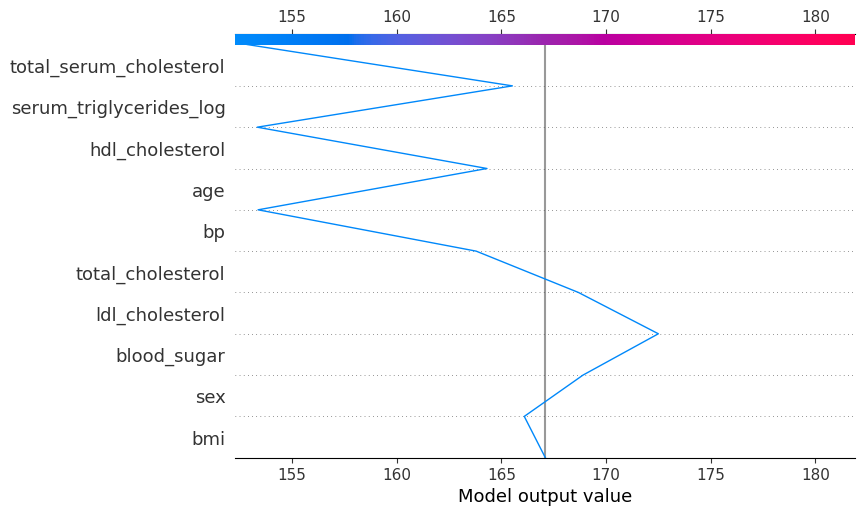
shap.plots.decision(
explainer.expected_value,
explainer.shap_values(diabetes_X_test)[0],
feature_names=X.columns.tolist()
)
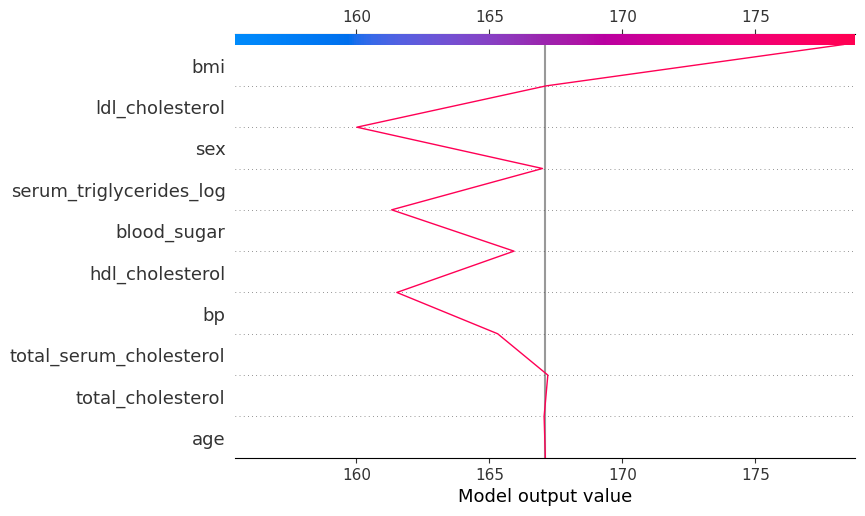
shap.plots.decision(
explainer.expected_value,
explainer.shap_values(diabetes_X_test)[104],
feature_names=X.columns.tolist()
)
17.8 Group Difference
bmi_category_obese = np.array([False if shap_values[i, "bmi"].data < 30 else True for i in range(shap_values.shape[0])])
# bmi_category_obese
shap.plots.group_difference(shap_values_numeric, bmi_category_obese, feature_names=X.columns.tolist())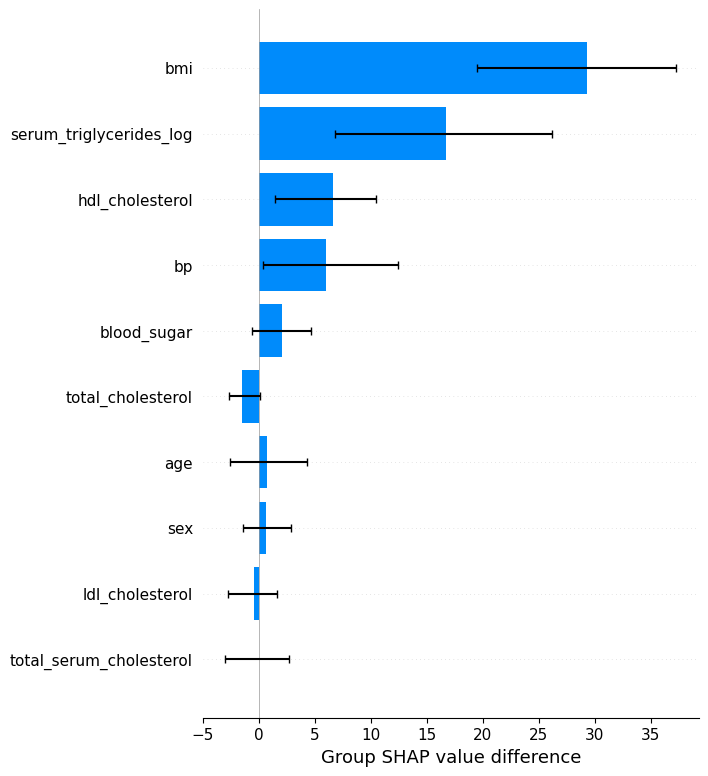
17.9 Modifying plots
shap.plots.decision(
explainer.expected_value,
explainer.shap_values(X)[125],
feature_names=X.columns.tolist(),
show=False ## NEW
)
plt.gcf() ## NEW
ax = plt.title("Here is my added title") ## NEW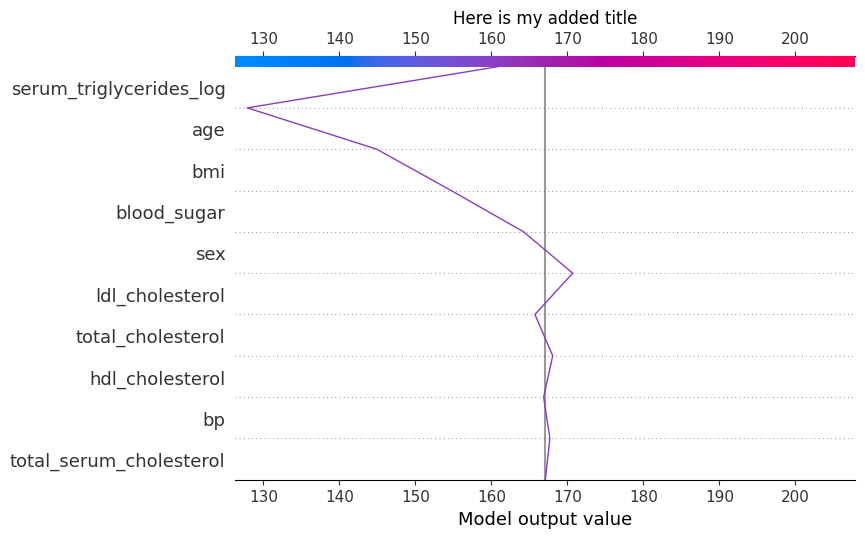
17.10 Subplots
fig, (ax1, ax2) = plt.subplots(nrows=1, ncols=2, figsize=(15,10)) ## NEW
# sca is 'set current axis'
# ensures next plot is put onto the axis we specify here - our first
# of the two subplots
plt.sca(ax1) ## NEW
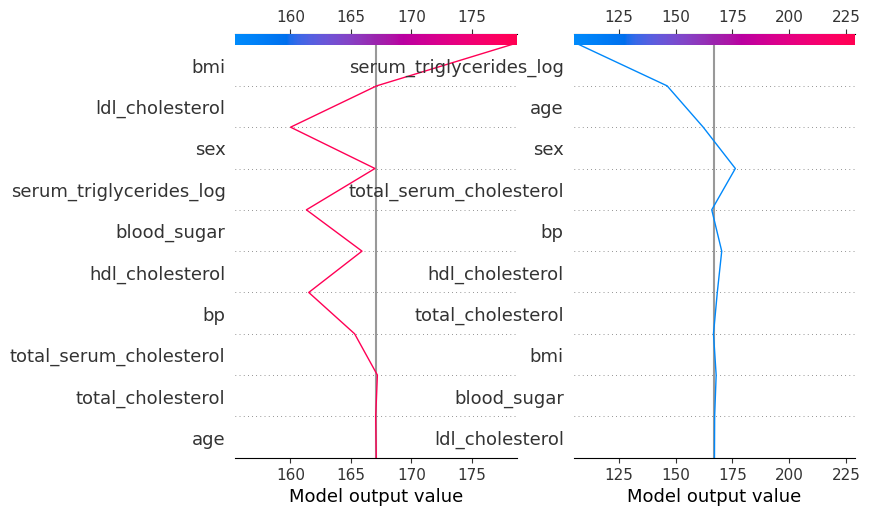
shap.plots.decision(
explainer.expected_value,
explainer.shap_values(diabetes_X_test)[104],
feature_names=X.columns.tolist(),
show=False ## NEW
)
# Change to the second axis
plt.sca(ax2) ## NEW
shap.plots.decision(
explainer.expected_value,
explainer.shap_values(diabetes_X_test)[15],
feature_names=X.columns.tolist(),
show=False ## NEW
)
# note that the use of %matplotlib inline at the start has led to the figsize parameter being
# partly ignored