import pandas as pd
import numpy as np
# Download data
# (not required if running locally and have previously downloaded data)
download_required = True
if download_required:
# Download processed data:
address = 'https://raw.githubusercontent.com/MichaelAllen1966/' + \
'2004_titanic/master/jupyter_notebooks/data/hsma_stroke.csv'
data = pd.read_csv(address)
# Create a data subfolder if one does not already exist
import os
data_directory ='./data/'
if not os.path.exists(data_directory):
os.makedirs(data_directory)
# Save data to data subfolder
data.to_csv(data_directory + 'hsma_stroke.csv', index=False)
# Load data
data = pd.read_csv('data/hsma_stroke.csv')
# Make all data 'float' type
data = data.astype(float)19 Exercise Solution: Explainable AI (Stroke Thromobolysis Dataset)
The data loaded in this exercise is for seven acute stroke units, and whether a patient receives clost-busting treatment for stroke. There are lots of features, and a description of the features can be found in the file stroke_data_feature_descriptions.csv.
Train a decision tree model to try to predict whether or not a stroke patient receives clot-busting treatment. Use the prompts below to write each section of code.
Run the code below to import the dataset.
Import the libraries you may need! What was used last time?
What additional libraries might you need to import to try out the different boosted trees?
# Import machine learning methods
from xgboost.sklearn import XGBClassifier
from sklearn.model_selection import train_test_split
from sklearn.tree import plot_tree
import plotly.express as px
import matplotlib.pyplot as plt
from sklearn.metrics import auc, roc_curve, RocCurveDisplay, f1_score, precision_score, \
recall_score, confusion_matrix, ConfusionMatrixDisplay, \
classification_report# Additional imports for explainable
from sklearn.inspection import PartialDependenceDisplay, permutation_importance
# Import shap for shapley values
import shap
# JavaScript Important for the interactive charts later on
shap.initjs()X = data.drop('Clotbuster given',axis=1) # X = all 'data' except the 'survived' column
y = data['Clotbuster given'] # y = 'survived' column from 'data'
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.25, random_state=42)
model = XGBClassifier(random_state=42)
model = model.fit(X_train,y_train)
# Predict training and test set labels
y_pred_train = model.predict(X_train)
y_pred_test = model.predict(X_test)
accuracy_train = np.mean(y_pred_train == y_train)
accuracy_test = np.mean(y_pred_test == y_test)
print (f'Accuracy of predicting training data = {accuracy_train}')
print (f'Accuracy of predicting test data = {accuracy_test}')Accuracy of predicting training data = 0.9978510028653295
Accuracy of predicting test data = 0.815450643776824120 Feature Importance
20.0.1 MDI
features = list(X_train)
feature_importances_xgb = model.feature_importances_
importances_xgb = pd.DataFrame(index=features)
importances_xgb['importance_xgb'] = feature_importances_xgb
importances_xgb['rank_xgb'] = importances_xgb['importance_xgb'].rank(ascending=False).values
importances_xgb.sort_values('rank_xgb').head()| importance_xgb | rank_xgb | |
|---|---|---|
| Stroke Type_I | 0.292050 | 1.0 |
| Onset Time Known Type_BE | 0.060007 | 2.0 |
| S2NihssArrival | 0.047182 | 3.0 |
| Anticoag before stroke_1 | 0.042693 | 4.0 |
| S2RankinBeforeStroke | 0.038013 | 5.0 |
20.0.2 PFI
feature_names = X.columns.tolist()
result_dt_pfi = permutation_importance(
model, X_test, y_test, n_repeats=10, random_state=42, n_jobs=2
)
importances_pfi_dt = pd.Series(result_dt_pfi.importances_mean, index=feature_names)
fig, ax = plt.subplots(figsize=(15,10))
importances_pfi_dt.plot.bar(yerr=result_dt_pfi.importances_std, ax=ax)
ax.set_title("Feature importances using permutation on full model")
ax.set_ylabel("Mean accuracy decrease")
fig.tight_layout()
plt.show()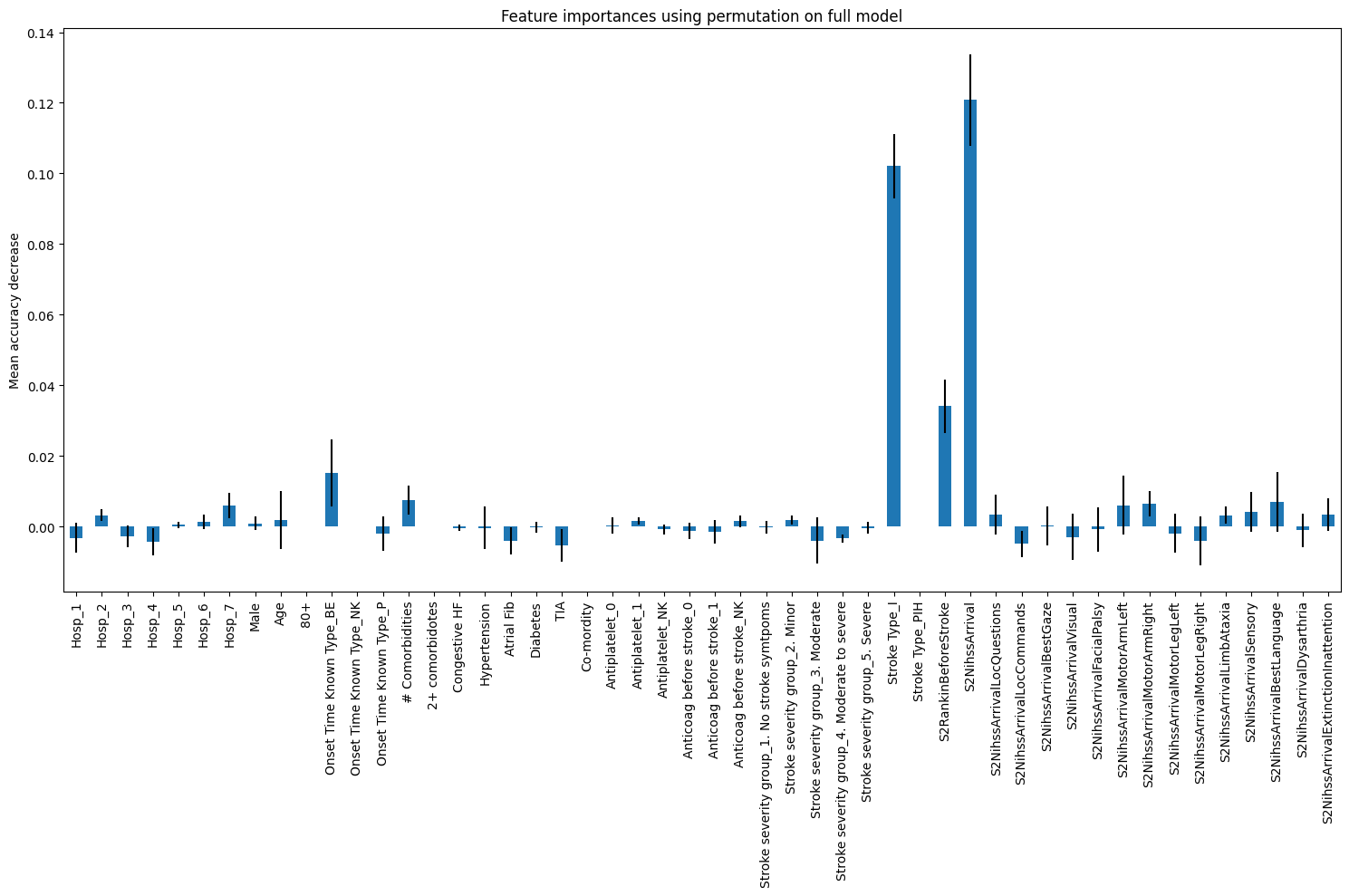
21 PDP + ICE
fig, ax = plt.subplots(figsize=(10, 6))
display = PartialDependenceDisplay.from_estimator(
model, # Your fitted model
X_train, # Your feature matrix
features=['Age'], # List of features to plot
target=0,
kind='individual', # Type of PDP
ax=ax,
random_state=42
)
plt.show()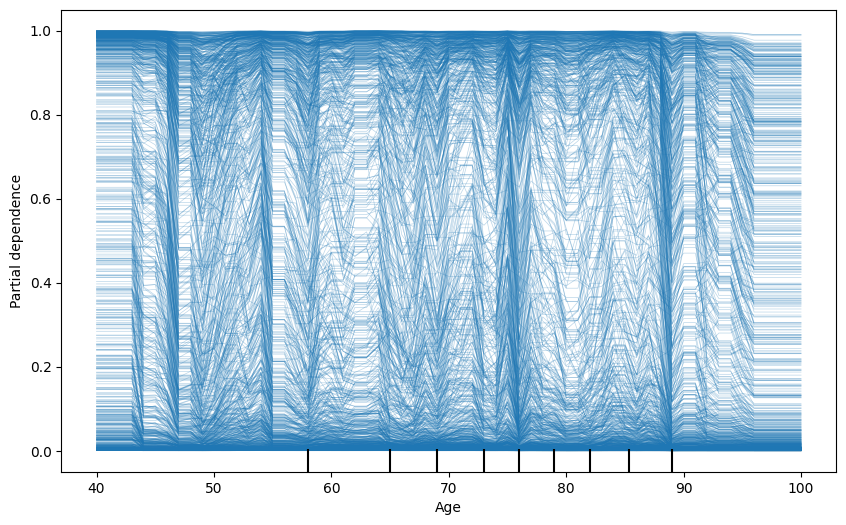
fig, ax = plt.subplots(figsize=(10, 6))
display = PartialDependenceDisplay.from_estimator(
model, # Your fitted model
X_train, # Your feature matrix
features=['Age'], # List of features to plot
target=0,
kind='both', # Type of PDP
ax=ax,
random_state=42
)
plt.show()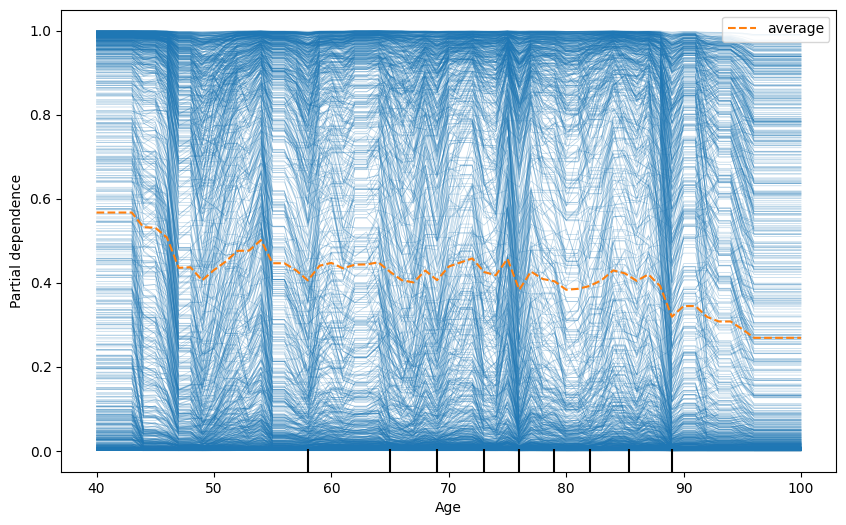
fig, ax = plt.subplots(figsize=(10, 6))
display = PartialDependenceDisplay.from_estimator(
model, # Your fitted model
X_train, # Your feature matrix
features=['Age'], # List of features to plot
target=0,
kind='average', # Type of PDP
ax=ax,
random_state=42
)
plt.show()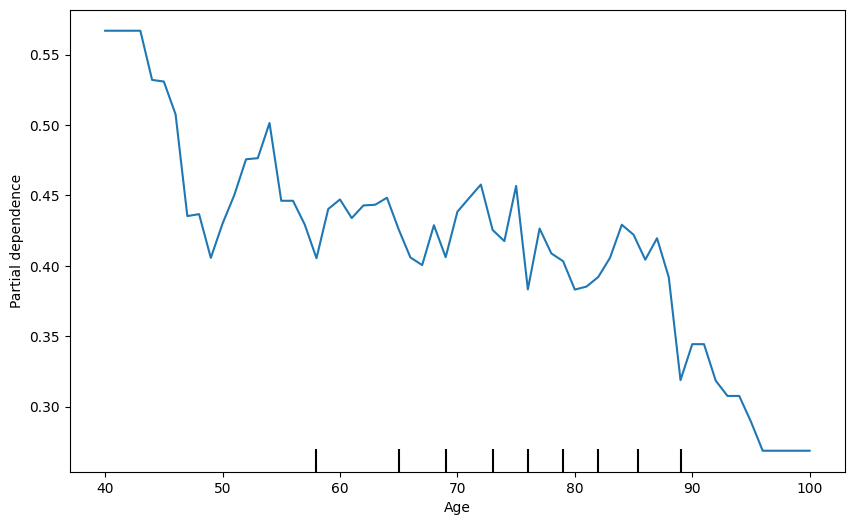
22 SHAP
# explain the model's predictions using SHAP
explainer = shap.Explainer(model, X_train)
shap_values = explainer(X_test)
shap_values.values =
array([[ 0.57191953, -0.00599206, -0.02803201, ..., 0.53735721,
-0.11320076, -0.18555162],
[ 0.01019792, -0.01376853, -0.20353487, ..., 0.0578289 ,
0.08007584, -0.10086989],
[-0.06371776, -0.05613441, 0.00877733, ..., -0.40052104,
-0.15933883, -0.24292226],
...,
[-0.00602321, 0.17367847, 0.0584153 , ..., -0.00764367,
0.18520108, -0.16023535],
[-0.02695623, -0.01631852, 0.035347 , ..., 0.75401062,
0.12778105, -0.06493782],
[-0.08913676, -0.06648799, 0.06200275, ..., -0.47741059,
-0.15630577, -0.15140597]])
.base_values =
array([-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877, -1.97564877, -1.97564877, -1.97564877, -1.97564877,
-1.97564877])
.data =
array([[1., 0., 0., ..., 2., 0., 0.],
[0., 0., 1., ..., 1., 0., 0.],
[0., 0., 1., ..., 0., 0., 0.],
...,
[0., 1., 0., ..., 1., 1., 0.],
[0., 0., 0., ..., 2., 1., 1.],
[0., 0., 0., ..., 0., 0., 0.]])shap_values_numeric = shap_values.values
shap_values_numericarray([[ 0.57191953, -0.00599206, -0.02803201, ..., 0.53735721,
-0.11320076, -0.18555162],
[ 0.01019792, -0.01376853, -0.20353487, ..., 0.0578289 ,
0.08007584, -0.10086989],
[-0.06371776, -0.05613441, 0.00877733, ..., -0.40052104,
-0.15933883, -0.24292226],
...,
[-0.00602321, 0.17367847, 0.0584153 , ..., -0.00764367,
0.18520108, -0.16023535],
[-0.02695623, -0.01631852, 0.035347 , ..., 0.75401062,
0.12778105, -0.06493782],
[-0.08913676, -0.06648799, 0.06200275, ..., -0.47741059,
-0.15630577, -0.15140597]])22.0.1 Feature table
# get feature importance for comparison using MDI method
features = list(X_train)
feature_importances = model.feature_importances_
importances = pd.DataFrame(index=features)
importances['importance'] = feature_importances
importances['rank'] = importances['importance'].rank(ascending=False).values
importances.sort_values('rank').head()
# Get shapley importances
# Calculate mean Shapley value for each feature in trainign set
importances['mean_shapley_values'] = np.mean(
shap_values_numeric, axis=0
)
# Calculate mean absolute Shapley value for each feature in trainign set
# This will give us the average importance of each feature
importances['mean_abs_shapley_values'] = np.mean(
np.abs(shap_values_numeric), axis=0
)
importances| importance | rank | mean_shapley_values | mean_abs_shapley_values | |
|---|---|---|---|---|
| Hosp_1 | 0.016272 | 14.0 | 0.048697 | 0.113734 |
| Hosp_2 | 0.009090 | 41.0 | 0.007687 | 0.070485 |
| Hosp_3 | 0.011619 | 30.0 | 0.009452 | 0.054508 |
| Hosp_4 | 0.025572 | 6.0 | -0.006476 | 0.071526 |
| Hosp_5 | 0.016513 | 13.0 | 0.000117 | 0.025848 |
| Hosp_6 | 0.018912 | 8.0 | -0.017381 | 0.070901 |
| Hosp_7 | 0.009989 | 37.0 | 0.020218 | 0.148842 |
| Male | 0.011162 | 32.0 | 0.011670 | 0.089067 |
| Age | 0.012259 | 28.0 | 0.095522 | 0.513481 |
| 80+ | 0.000000 | 48.0 | 0.000000 | 0.000000 |
| Onset Time Known Type_BE | 0.060007 | 2.0 | -0.035751 | 0.513144 |
| Onset Time Known Type_NK | 0.000000 | 48.0 | 0.000000 | 0.000000 |
| Onset Time Known Type_P | 0.021683 | 7.0 | -0.012942 | 0.105682 |
| # Comorbidities | 0.012480 | 27.0 | 0.008446 | 0.140819 |
| 2+ comorbidotes | 0.000000 | 48.0 | 0.000000 | 0.000000 |
| Congestive HF | 0.008597 | 42.0 | -0.004122 | 0.008599 |
| Hypertension | 0.012239 | 29.0 | -0.070380 | 0.250935 |
| Atrial Fib | 0.016634 | 12.0 | 0.011129 | 0.097199 |
| Diabetes | 0.017148 | 11.0 | -0.002177 | 0.020622 |
| TIA | 0.016100 | 15.0 | -0.013201 | 0.363766 |
| Co-mordity | 0.000000 | 48.0 | 0.000000 | 0.000000 |
| Antiplatelet_0 | 0.009424 | 39.0 | 0.002172 | 0.017998 |
| Antiplatelet_1 | 0.006572 | 43.0 | -0.001211 | 0.024097 |
| Antiplatelet_NK | 0.013325 | 23.0 | 0.028037 | 0.113407 |
| Anticoag before stroke_0 | 0.014763 | 17.0 | -0.055988 | 0.227291 |
| Anticoag before stroke_1 | 0.042693 | 4.0 | 0.023494 | 0.148024 |
| Anticoag before stroke_NK | 0.014519 | 18.0 | -0.002497 | 0.019437 |
| Stroke severity group_1. No stroke symtpoms | 0.004698 | 44.0 | 0.003031 | 0.014462 |
| Stroke severity group_2. Minor | 0.002441 | 45.0 | -0.000203 | 0.020181 |
| Stroke severity group_3. Moderate | 0.009636 | 38.0 | 0.015730 | 0.187784 |
| Stroke severity group_4. Moderate to severe | 0.010140 | 36.0 | -0.000302 | 0.010772 |
| Stroke severity group_5. Severe | 0.011573 | 31.0 | -0.003786 | 0.015024 |
| Stroke Type_I | 0.292050 | 1.0 | 0.263943 | 1.526787 |
| Stroke Type_PIH | 0.000000 | 48.0 | 0.000000 | 0.000000 |
| S2RankinBeforeStroke | 0.038013 | 5.0 | -0.010677 | 0.460929 |
| S2NihssArrival | 0.047182 | 3.0 | 0.167792 | 1.423348 |
| S2NihssArrivalLocQuestions | 0.013432 | 22.0 | -0.009531 | 0.252463 |
| S2NihssArrivalLocCommands | 0.013001 | 25.0 | -0.004716 | 0.189615 |
| S2NihssArrivalBestGaze | 0.017906 | 10.0 | -0.049027 | 0.294514 |
| S2NihssArrivalVisual | 0.010915 | 34.0 | 0.016386 | 0.203184 |
| S2NihssArrivalFacialPalsy | 0.013921 | 19.0 | 0.044696 | 0.199442 |
| S2NihssArrivalMotorArmLeft | 0.018599 | 9.0 | -0.028893 | 0.237647 |
| S2NihssArrivalMotorArmRight | 0.009192 | 40.0 | 0.018928 | 0.069969 |
| S2NihssArrivalMotorLegLeft | 0.011116 | 33.0 | 0.020311 | 0.108674 |
| S2NihssArrivalMotorLegRight | 0.013778 | 21.0 | -0.002636 | 0.231136 |
| S2NihssArrivalLimbAtaxia | 0.010385 | 35.0 | -0.008956 | 0.038973 |
| S2NihssArrivalSensory | 0.012529 | 26.0 | -0.020345 | 0.162434 |
| S2NihssArrivalBestLanguage | 0.013232 | 24.0 | -0.006986 | 0.421014 |
| S2NihssArrivalDysarthria | 0.014873 | 16.0 | -0.016975 | 0.148263 |
| S2NihssArrivalExtinctionInattention | 0.013818 | 20.0 | 0.011012 | 0.232744 |
22.0.1.1 Feature importance comparison
importance_top_10 = \
importances.sort_values(
by='importance', ascending=False
).head(10).index
shapley_top_10 = \
importances.sort_values(
by='mean_abs_shapley_values',
ascending=False).head(10).index
# Add to DataFrame
top_10_features = pd.DataFrame()
top_10_features['importances'] = importance_top_10.values
top_10_features['Shapley'] = shapley_top_10.values
# Display
top_10_features| importances | Shapley | |
|---|---|---|
| 0 | Stroke Type_I | Stroke Type_I |
| 1 | Onset Time Known Type_BE | S2NihssArrival |
| 2 | S2NihssArrival | Age |
| 3 | Anticoag before stroke_1 | Onset Time Known Type_BE |
| 4 | S2RankinBeforeStroke | S2RankinBeforeStroke |
| 5 | Hosp_4 | S2NihssArrivalBestLanguage |
| 6 | Onset Time Known Type_P | TIA |
| 7 | Hosp_6 | S2NihssArrivalBestGaze |
| 8 | S2NihssArrivalMotorArmLeft | S2NihssArrivalLocQuestions |
| 9 | S2NihssArrivalBestGaze | Hypertension |
22.0.2 Global: Beeswarm
# summarize the effects of all the features
shap.plots.beeswarm(shap_values, max_display=25)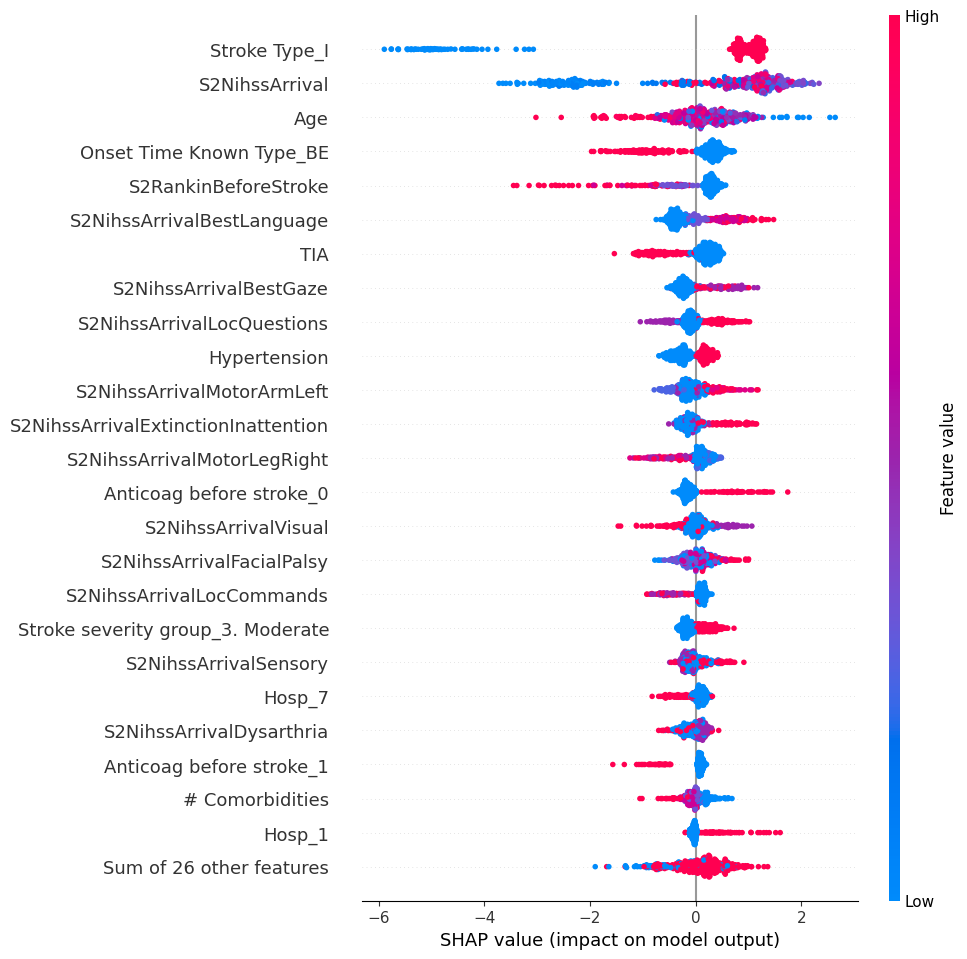
22.0.3 Global: Bar
shap.plots.bar(shap_values, max_display=20)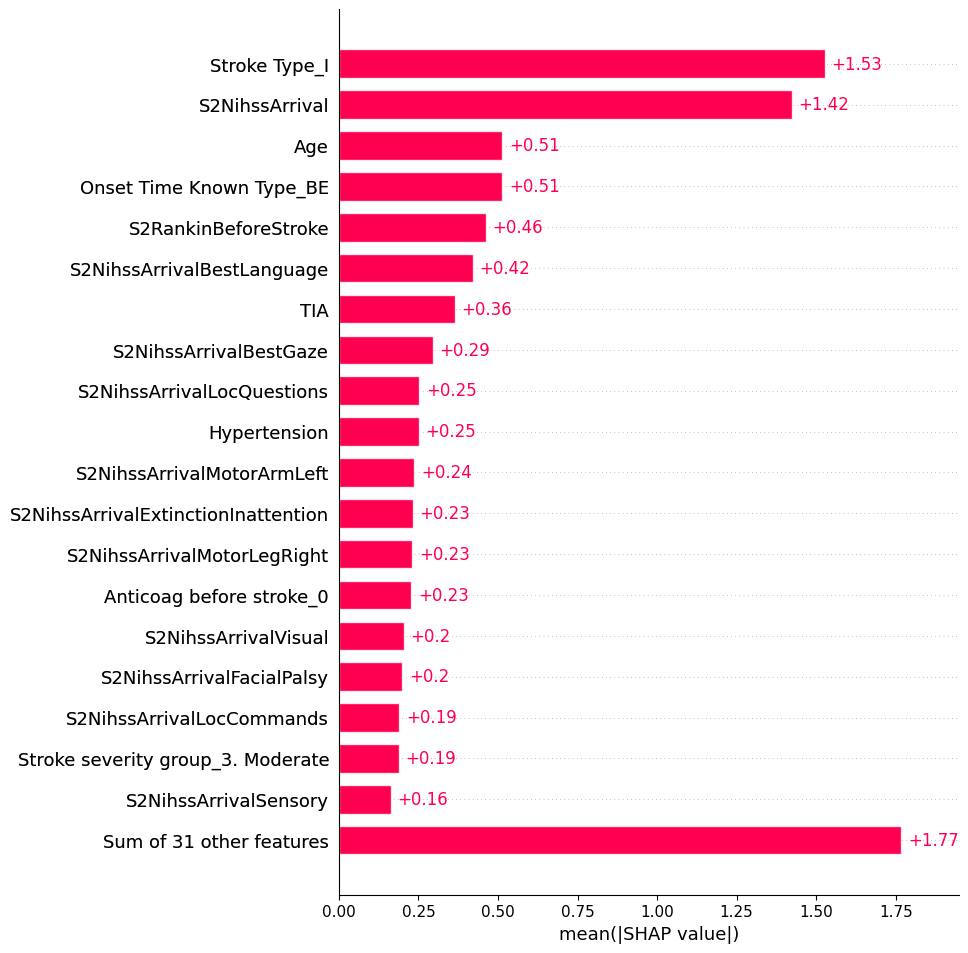
22.0.4 Bar: by factor
sex = ["Women" if shap_values[i, "Male"].data == 0 else "Men" for i in range(shap_values.shape[0])]
shap.plots.bar(shap_values.cohorts(sex).abs.mean(0))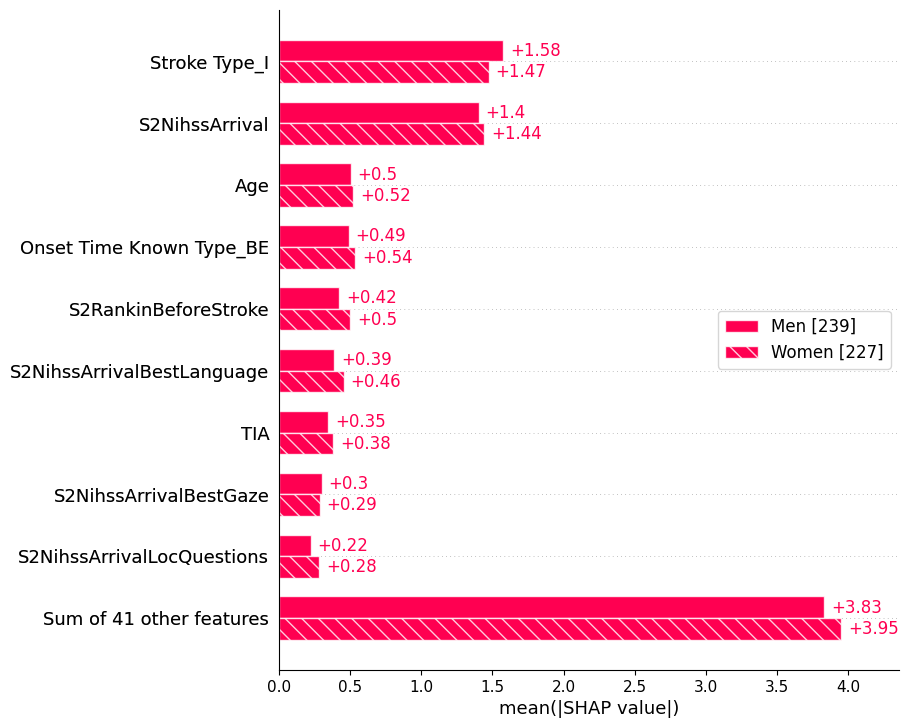
22.1 Local: Waterfall plots
# visualize the first prediction's explanation
shap.plots.waterfall(shap_values[0])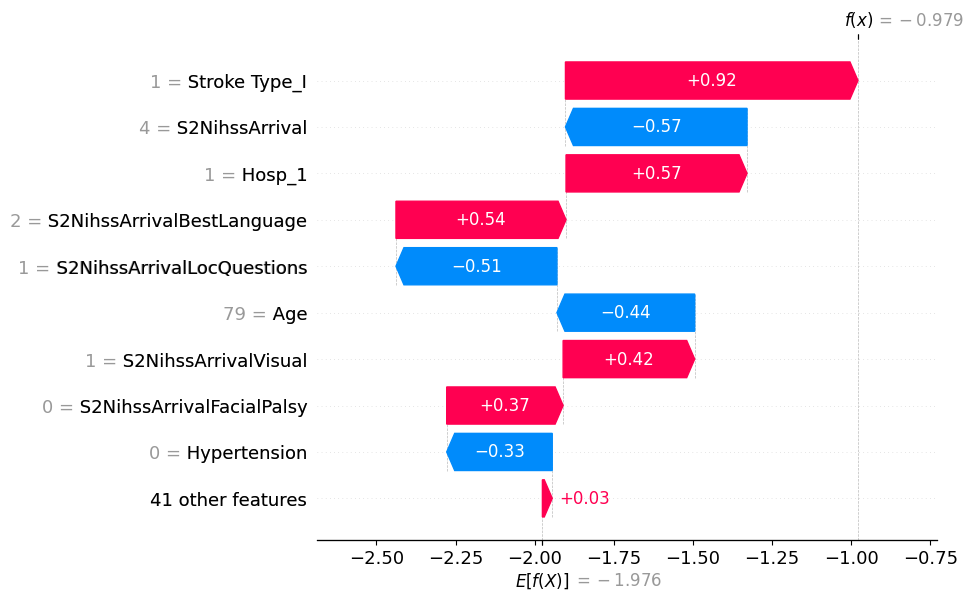
# visualize another prediction's explanation
shap.plots.waterfall(shap_values[7], max_display=15)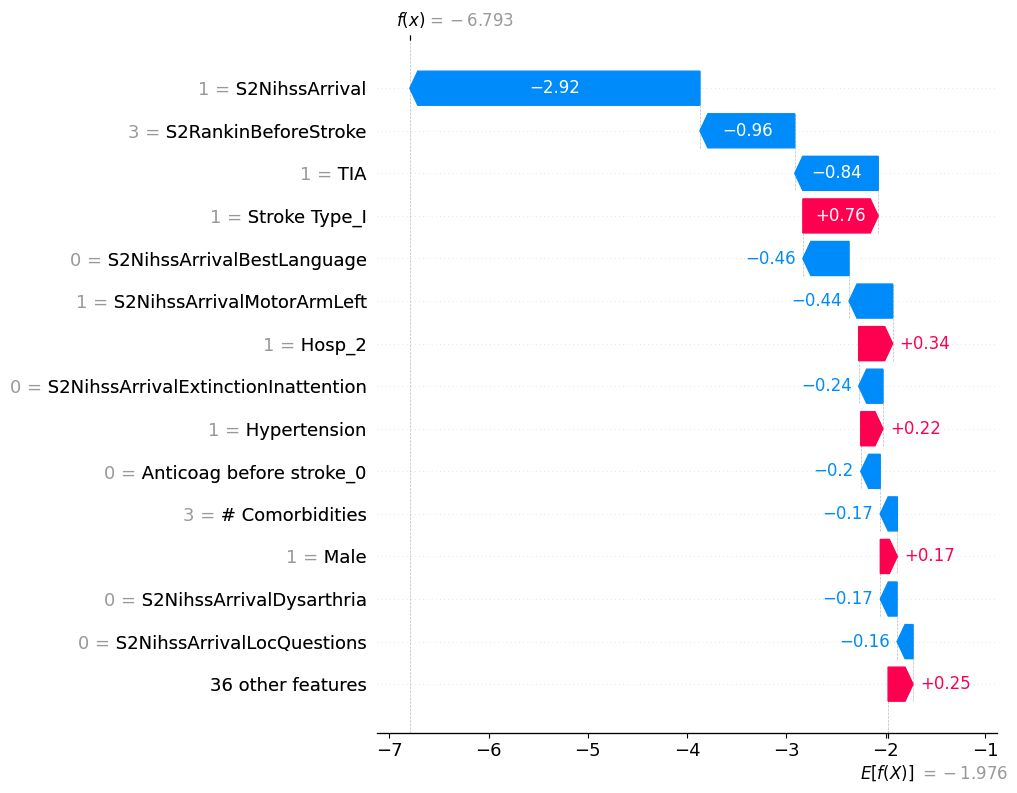
# visualize another prediction's explanation
shap.plots.waterfall(shap_values[145], max_display=15)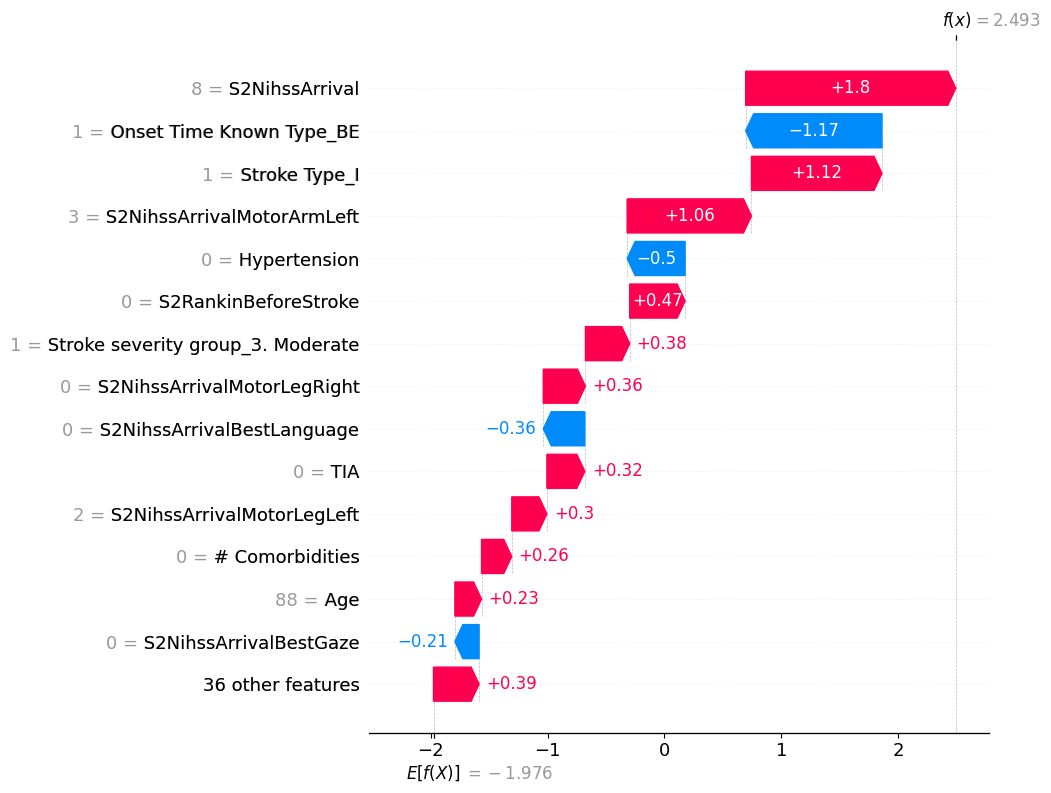
22.1.1 Local: Force Plots
# visualize the first prediction's explanation with a force plot
shap.plots.force(shap_values[0])Have you run `initjs()` in this notebook? If this notebook was from another user you must also trust this notebook (File -> Trust notebook). If you are viewing this notebook on github the Javascript has been stripped for security. If you are using JupyterLab this error is because a JupyterLab extension has not yet been written.
22.1.2 Global: Force Plots
# visualize all the predictions
# this struggles with a large number of values so we'll sample a small set
shap.plots.force(shap.utils.sample(shap_values, 1000))Have you run `initjs()` in this notebook? If this notebook was from another user you must also trust this notebook (File -> Trust notebook). If you are viewing this notebook on github the Javascript has been stripped for security. If you are using JupyterLab this error is because a JupyterLab extension has not yet been written.
22.1.3 Dependence Plots
X.columnsIndex(['Hosp_1', 'Hosp_2', 'Hosp_3', 'Hosp_4', 'Hosp_5', 'Hosp_6', 'Hosp_7',
'Male', 'Age', '80+', 'Onset Time Known Type_BE',
'Onset Time Known Type_NK', 'Onset Time Known Type_P',
'# Comorbidities', '2+ comorbidotes', 'Congestive HF', 'Hypertension',
'Atrial Fib', 'Diabetes', 'TIA', 'Co-mordity', 'Antiplatelet_0',
'Antiplatelet_1', 'Antiplatelet_NK', 'Anticoag before stroke_0',
'Anticoag before stroke_1', 'Anticoag before stroke_NK',
'Stroke severity group_1. No stroke symtpoms',
'Stroke severity group_2. Minor', 'Stroke severity group_3. Moderate',
'Stroke severity group_4. Moderate to severe',
'Stroke severity group_5. Severe', 'Stroke Type_I', 'Stroke Type_PIH',
'S2RankinBeforeStroke', 'S2NihssArrival', 'S2NihssArrivalLocQuestions',
'S2NihssArrivalLocCommands', 'S2NihssArrivalBestGaze',
'S2NihssArrivalVisual', 'S2NihssArrivalFacialPalsy',
'S2NihssArrivalMotorArmLeft', 'S2NihssArrivalMotorArmRight',
'S2NihssArrivalMotorLegLeft', 'S2NihssArrivalMotorLegRight',
'S2NihssArrivalLimbAtaxia', 'S2NihssArrivalSensory',
'S2NihssArrivalBestLanguage', 'S2NihssArrivalDysarthria',
'S2NihssArrivalExtinctionInattention'],
dtype='object')22.1.3.1 Simple scatter of a single feature
# create a dependence scatter plot to show the effect of a single feature across the whole dataset
shap.plots.scatter(shap_values[:, 'Age'])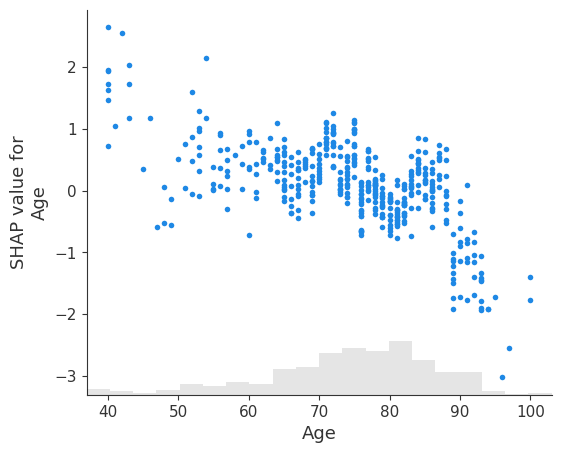
shap.plots.scatter(shap_values[:, 'Diabetes'])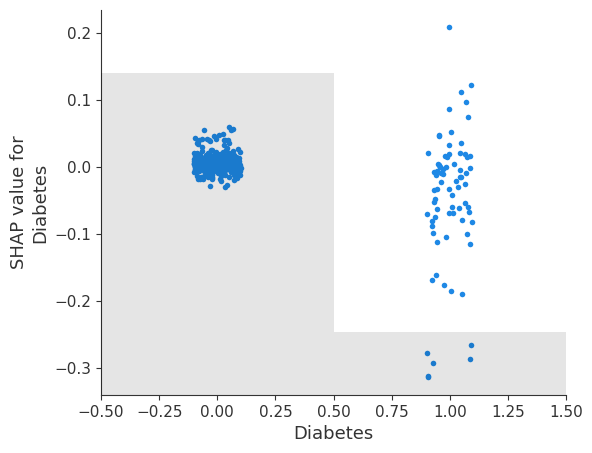
shap.plots.scatter(shap_values[:, '# Comorbidities'])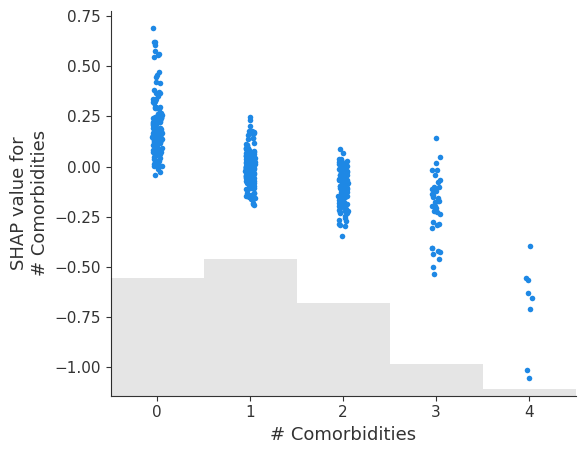
22.1.4 Scatter of multiple features
Passing in shap_values to the colour will colour the value by the most strongly interacting other value.
# create a dependence scatter plot to show the effect of a single feature across the whole dataset
shap.plots.scatter(shap_values[:, "Male"], color=shap_values)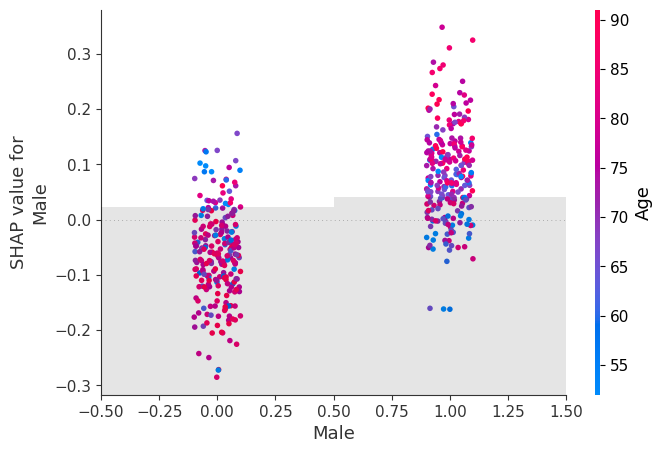
# create a dependence scatter plot to show the effect of a single feature across the whole dataset
shap.plots.scatter(shap_values[:, "Age"], color=shap_values)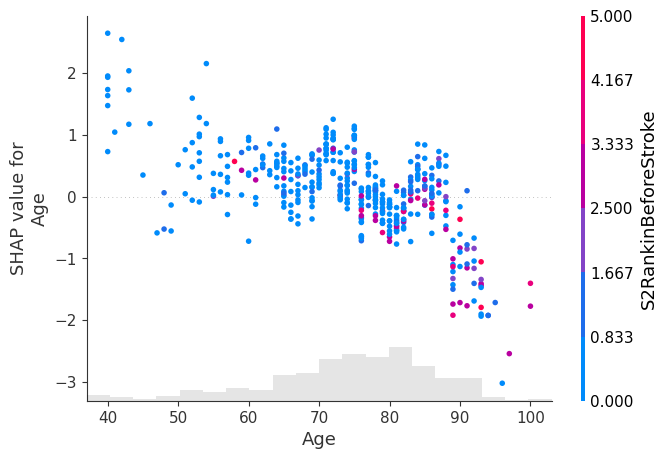
Alternatively we can choose to colour by a specific column.
# create a dependence scatter plot to show the effect of a single feature across the whole dataset
shap.plots.scatter(shap_values[:, "Age"], color=shap_values[:,"Male"])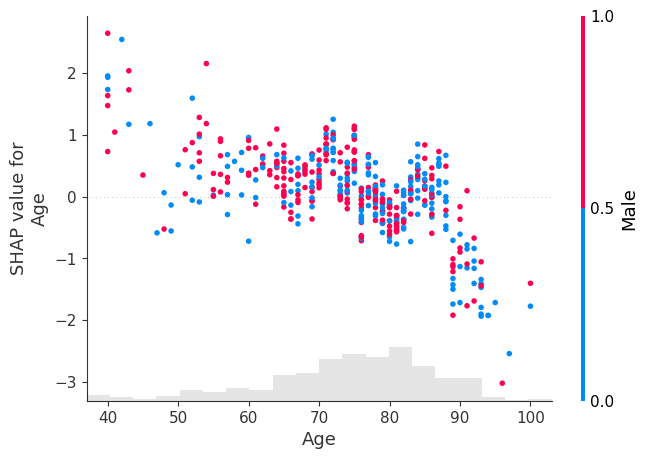
# create a dependence scatter plot to show the effect of a single feature across the whole dataset
shap.plots.scatter(shap_values[:, "Male"], color=shap_values[:,"Age"])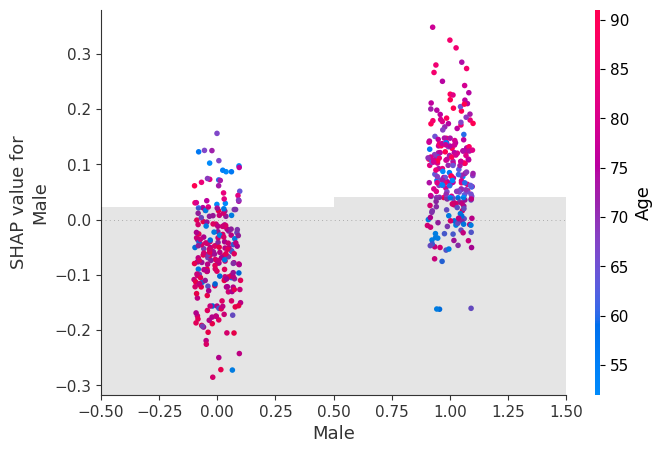
22.2 Prediction Uncertainty
splits = 50
train_set = []
for i in range(splits):
train_set.append(X_train.join(y_train).sample(frac=1, replace=True))# Set up lists for models and probability predictions
models = []
results = []
accuracies = []
for i in range(splits):
# Get X and y
X_train = train_set[i].drop('Clotbuster given', axis=1)
y_train = train_set[i]['Clotbuster given']
# Define and train model; use different random seed for each model
model = XGBClassifier(random_state=42+i)
model.fit(X_train, y_train)
models.append(model)
# Get predicted probabilities and class
y_probs = model.predict_proba(X_test)[:,1]
y_pred = y_probs > 0.5
results.append([y_probs])
# Show accuracy
accuracy = np.mean(y_pred == y_test)
accuracies.append(accuracy)
results = np.array(results)
results = results.T.reshape(-1, splits)print (f'Mean accuracy: {np.mean(accuracies):0.3f}')Mean accuracy: 0.803fig = plt.figure()
ax = fig.add_subplot()
ax.boxplot(accuracies, whis=999)
ax.set_ylabel('Model accuracy')
ax.axes.xaxis.set_ticklabels([]) # Remove xtick labels
plt.show()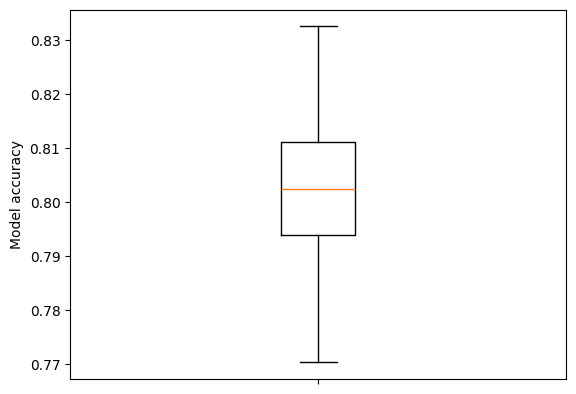
classification = results >= 0.5
consensus = classification.sum(axis=1) >= splits/2
consensus_accuracy = np.mean(consensus == y_test)
print (f'Consensus accuracy: {consensus_accuracy:0.3f}')Consensus accuracy: 0.826results = results[np.mean(results,axis=1).argsort()]
mean = np.mean(results,axis=1)
stdev = np.std(results,axis=1)
fig = plt.figure()
ax = fig.add_subplot()
ax.errorbar(range(len(mean)), mean, yerr=stdev, label='Standard Deviation', zorder=1)
ax.plot(mean, 'o', c='r', markersize=2, label = 'Mean probability', zorder=2)
ax.axes.xaxis.set_ticklabels([])
ax.set_xlabel('Patient')
ax.set_ylabel('Probability of Clotbuster Being Given')
ax.set_xticks([])
ax.grid()
ax.legend()
plt.show()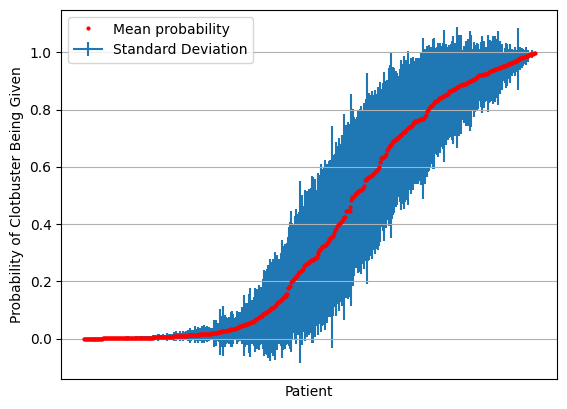
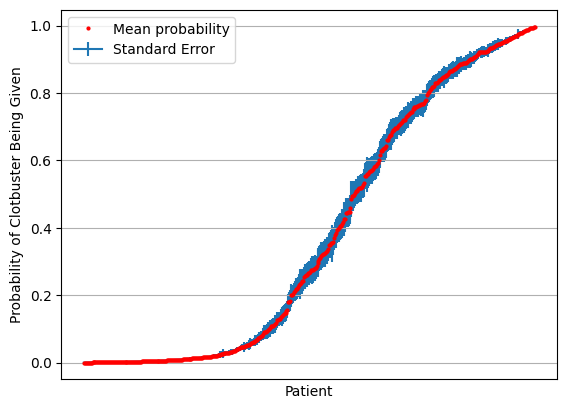
results = results[np.mean(results,axis=1).argsort()]
mean = np.mean(results,axis=1)
stdev = np.std(results,axis=1)
se = stdev / np.sqrt(splits)
fig = plt.figure()
ax = fig.add_subplot()
ax.errorbar(range(len(mean)), mean, yerr=se, label='Standard Error', zorder=1)
ax.plot(mean, 'o', c='r', markersize=2, label = 'Mean probability', zorder=2)
ax.axes.xaxis.set_ticklabels([])
ax.set_xlabel('Patient')
ax.set_ylabel('Probability of Clotbuster Being Given')
ax.set_xticks([])
ax.grid()
ax.legend()
plt.show()